Tristan Cragnolini
Post-doctoral researcher
2018-2021 Topf and Thalassinos groups , Birbeck, University of London, London.
Cryo-EM data modelling, mass spectrometry data analysis.
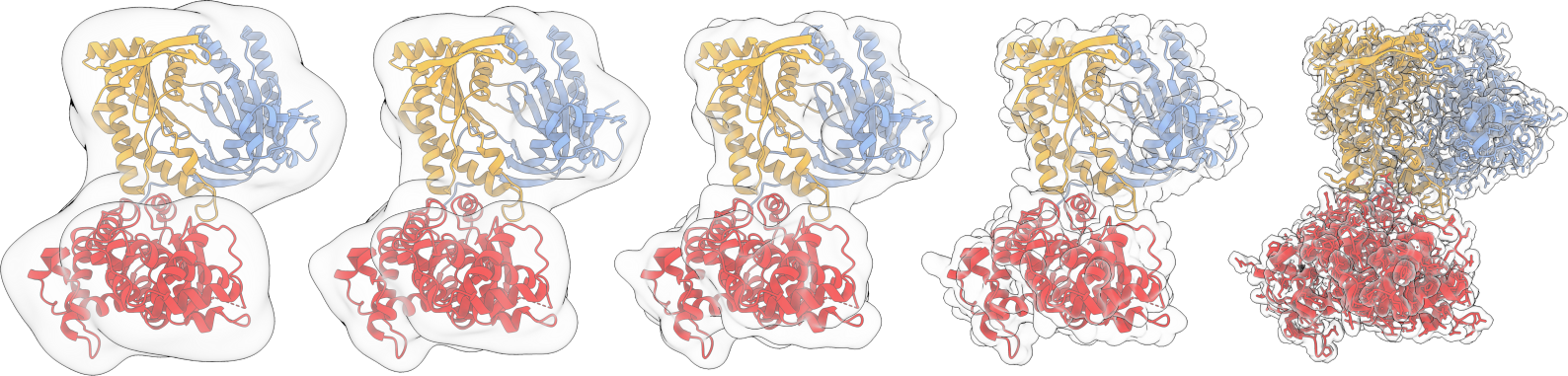
Rigid and flexible fitting of molecular model in cryo-EM maps. Development of statistical models for interpreting Ion Moblity-Mass Spectrometry data of protein assemblies.2014-2016 Wales group,University of Cambridge, Cambridge
Path sampling of coarse-grained biomolecules.
+Path sampling of G-quadruplex conformational transitions.+Monte-carlo simulations of RNA and protein folding.
+Benchmarking of RNA and protein forcefields
- September 2021, Invited Speaker, S2C2 CryoEM Map Modeling and Validation Workshop, Stanford, Online. Cragnolini T., Topf M., “Improving and validating map/model fits with TEMPy”.
- August 2021, Speaker, New directions of AI in structural biology, CIRM. §Cragnolini T., Kryshtafovych A., Topf M., “ML for cryo-EM”.
- December 2020, Speaker, CASP 14, Online. Cragnolini T., Topf M., Kryshtafovych A., “Evaluating and refining EM structures with TEMPy”.
- June 2020 Speaker, Annual Conference of the American Society for Mass Spectrometry, Online. Cragnolini T., Eldrid C., Brit H., Menneteau T., Ben-Younis A., Thalassinos K., “Automated, web-based analysis and visualisation of tandem ion mobility mass spectrometry data
- June 2019 Poster, Annual Conference of the American Society for Mass Spectrometry, Manchester, UK. Cragnolini T., Eldrid C., Brit H., Thalassinos K., “Automated, web-based analysis and visualisation of data from a Cyclic Ion Mobility Spectrometry Travelling Wave Device”.
- February 2014, Biophysical Society Meeting, San Francisco Poster: Cragnolini T., Sutherland-Cash K.H., Wales D., Pasquali S., Derreumaux P., “Wide exploration of OPEP protein energy landscapes using advanced Monte Carlo methods”.
- Poster: Cragnolini T., Laurin Y., Derreumaux P., Pasquali S., “Predicting RNA complex architectures”. May 2013, “Computational Biology: Then and Now” symposium, Weizmann Institute
- Poster: Cragnolini T., Derreumaux P., Pasquali S., “Multiscale modelling of RNAs combining coarse-grain folding with detailed all-atom dynamics”. April 2013, CFCAM Meeting, Paris, France
- Presentation: “Fully flexible coarse-grained model for RNA and DNA” January 2012, Amsterdam, Netherlands
- MOLSIM 2012 school: Understanding Molecular Simulation
- May 2011, groupe de graphisme et modélisation moléculaire (GGMM), La Rochelle Poster: Cragnolini T., Derreumaux P., Pasquali, S., “All-atom reconstruction of predicted coarse-grained RNA structures”
-
Post-doc Department of Chemistry, University of Cambridge
Supervisor: Lucy Colwell
RNA folding, detection of structural motifs - Post-doc Department of Chemistry, University of Cambridge10/2014 - 09/2016
Supervisor: David Wales
Path sampling of coarse-grained biomolecules - PhD thesis, Laboratoire de Biochimie Théorique, Paris, France10/2011 - 09/2014
Supervisors: Dr. Samuela Pasquali and Pr. Philippe Derreumaux
Predicting RNA structures and the large scale motions of RNA
using computer simulations and coarse-grained models.- Development of HiRE-RNA, a coarse-grain forcefield for RNA
- Optimization of the forcefield using a genetic algorithm
- Implementation of the forcefield in the OPEP package (Fortran)
- Development of analysis tools, in C++, Python and R
- All-atom RNA simulations with AMBER and Gromacs
- Masters intership, Computational Biology Unit, Bergen, Norway03/2010 - 06/2010
Supervisors: Dr. Nathalie Reuter and Svenn Grindhaug - Contribution to webnm@, a normal mode webserver for analysis and visualisation
Web address: http://apps.cbu.uib.no/webnma- Experience with MMTK, for the normal mode calculation (Python)
- Addition of a Jmol viewing applet, with visualisation of the modes (Javascript)
- Modification of the webserver to handle new analysis (Python, HTML, CSS, XML)
- Bachelor course, San Jose State University, San Jose, California, USA 03/2009 - 09/2009
Supervisor: Pr. Steve White - Work in a molecular biology laboratory (HeLa cell culture, Western Blot, PCR)
- 2011-2014 PhD thesis in molecular modelling, Université Paris Diderot-Paris 7, Paris, France
- 2009-2011 Master in bioinformatics, Université Paris Diderot-Paris 7, Paris, France
- 2008-2009 Exchange student at San José State University, San José, California, USA
- 2005-2009 Bachelor in chemistry and biology, Université Paris 12, Créteil, France
- 2015-2017 Supervision of classes in: computational techniques, quantum mechanics, perturbation theory, symmetry and bonding, University of Cambridge, UK
- 2011-2014 Teaching assistant for classes in Python(masters), bioinformatics(bachelor),
molecular modelling(masters), Université Paris Diderot-Paris 7, Paris, France - 2007-2008 Teaching assistant, Lycée Léon Blum (high school), Créteil, France
Molecular modelling Molecular modelling using Amber, GMIN, Gromacs, MMTK Models used: Amber99 (and variants), HiRE-RNA, Martini, OPEP Visualisation softwares: Chimera, Jmol, PyMol and VMD
Software development Programming languages : C, C++, Fortran(77/90), Python Data formatting using CSS, JSON, L AT EX, HTML, XML Parallel programming (MPI and pthreads), objected oriented programming Limited knowledge of Haskell, Java, Javascript, Perl, PHP, Development tools: Eclipse, Emacs, GDB, Git, SVN, Valgrind, Vi
Fluent English (2008, TOEFL: 105/120; 2011, Oxford placement test: level C2)
French (native language)
Beginner in Portuguese
Beginner in Spanish
This document was translated from L AT EX by H E V E A .