Protein and nucleic acid folding: disorder and ensembles
I have studied RNA, DNA and protein folding using different models. I have been particularly interested in metastable systems,
such as riboswitches, G-quadruplexes, and co-folding proteins.
I am keen on delving further into the assembly of larger biomolecular complexes.
To do that, accurate models of the smaller structural elements is required,
as well as new ways to coarse-grain those interactions in a way that allow for the modelling of large assemblies.
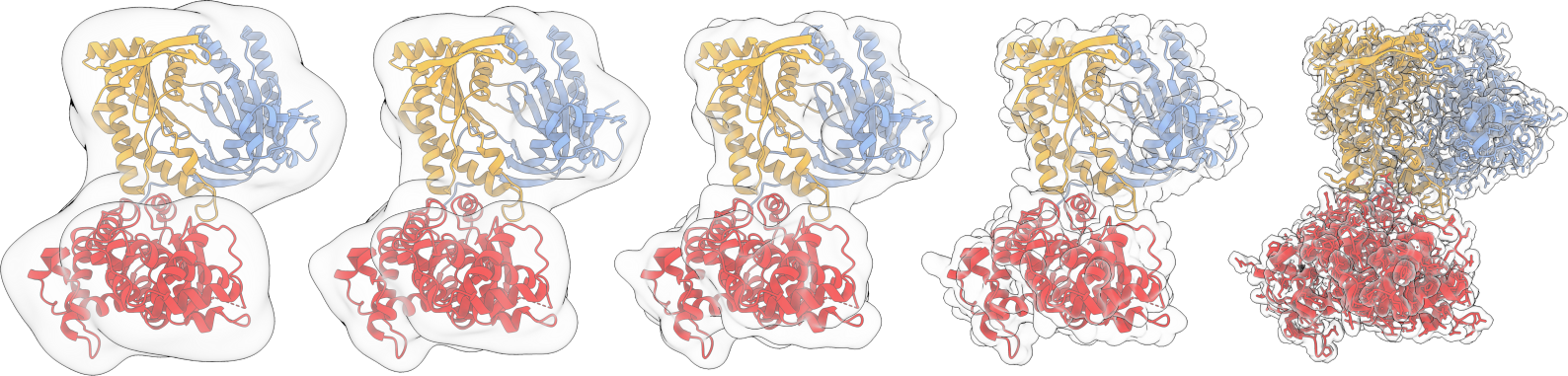
Coarse-grain model
With my PhD supervisor Samuela Pasquali, we worked on developing the HiRE-RNA forcefield to represent complex interactions in RNA and DNa,
and create a model that could truly represent the full range of possible RNA structures, without external constraints that do not correspond
to a biophysical force. This 'bottom-up' approach could then be combined with experimental restraints, as obtainable from cryo-EM, SAXS, etc...
I would like to extend this approach, and in particular work on automatising the process of developing these coarse-grained representation.
Biological assembly simulations
Work on G-quadruplexes with David Wales, Debayan Chakraborty, Liuba Mazzanti, Jiří Šponer, Samuela Pasquali, Philippe Derreumaux. We obtained pathways between common states using MD and pathsampling.
Model experimental setups
MS experimentsGraph enumeration of RNA structures
Work in collaboration with Ofer Kimchi, Lucy Colwell, and Michael Brenner.
SELEX analysis and simulation
Trying to understand how SELEX works, what can we tell about the obtained sequence, the likeliness of them binding to the target.
SELEX analysis
Work with Centauri (Helen Lavender, Alex Martin).
SELEX simulation
Work with Lucy Colwell, Alaa Abdel-Latif