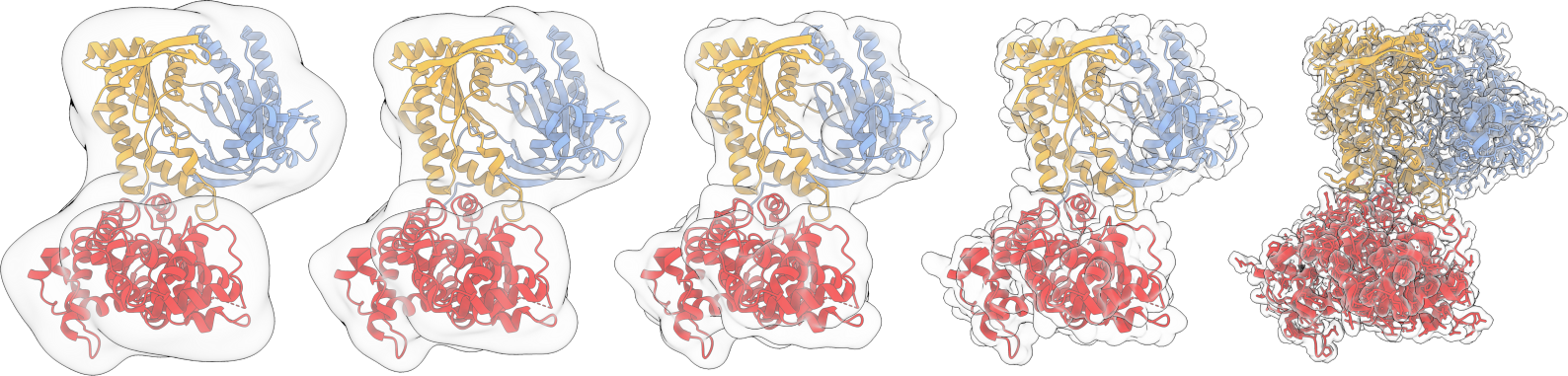
Flexible cryo-EM fitting
 A flexible fitting program, that uses GMM to estimate optimal positions and B-factors for each atom.
A flexible fitting program, that uses GMM to estimate optimal positions and B-factors for each atom.
TEMPy
 TEMPy is a software package to process cryo-EM maps, validate and fit structures to them.
TEMPy is a software package to process cryo-EM maps, validate and fit structures to them.
MS-UI
An analysis platform and web user interface that allows to combine and interpret IM-MS data, and slice IM-MS data.
G-quadruplex folding
Work with David Wales, Debayan Chakraborty, Liuba Mazzanti, Jiří Šponer, Samuela Pasquali, Philippe Derreumaux. We obtained pathways between common states using MD and pathsampling.
Graph enumeration of RNA structures
Work in collaboration with Ofer Kimchi, Lucy Colwell, and Michael Brenner.
HiRE-RNA coarse-grain model
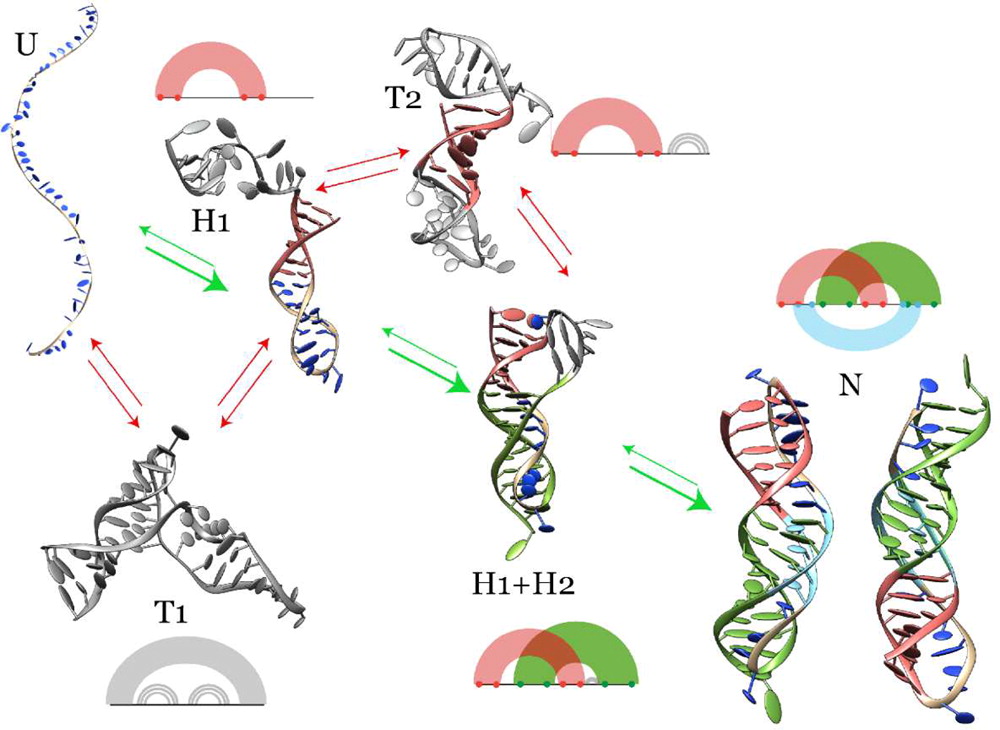 HiRE-RNA is a coarse grain nucleic acid forcefield, develop to accurately reproduce the interactions between nucleobases
that drives nucleic acid folding.
Work with Samuela Pasquali. Parametrisation with Yoann Laurin.
HiRE-RNA is a coarse grain nucleic acid forcefield, develop to accurately reproduce the interactions between nucleobases
that drives nucleic acid folding.
Work with Samuela Pasquali. Parametrisation with Yoann Laurin.
HiRE-RNA coarse-grain model
 HiRE-RNA is a coarse grain nucleic acid forcefield, develop to accurately reproduce the interactions between nucleobases
that drives nucleic acid folding.
Work with Samuela Pasquali. Parametrisation with Yoann Laurin.
HiRE-RNA is a coarse grain nucleic acid forcefield, develop to accurately reproduce the interactions between nucleobases
that drives nucleic acid folding.
Work with Samuela Pasquali. Parametrisation with Yoann Laurin.
Webnm@

INM
Interpolation with Network Model is a small program that computes an interpolation between two protein structures by moving along low-frequency normal modes computed on a structure, towards the final structure.